Rejection Sampling and Tricky Priors
This post is part of our Guide to Bayesian Statistics
At this point, we've covered the basics of Parameter Estimation, including building out the equivalent of a Hypothesis Test. We discussed using a Bayesian Prior when estimating parameters, but it turns out we've made some pretty big simplifying assumptions by always choosing to use a Beta distribution for our Prior. When assuming a Beta distribution for the Likelihood of our parameter it is often useful to assume that our Prior is also distributed according to a Beta distribution. This is primarily because it makes it very easy for us to combine the likelihood and prior analytically to derive our normalized Posterior probability distribution. However, this is quite a bit of a limitation since we need to have a pretty good guess as to what the peak of the distribution is.
Let's say you start working for an e-commerce company that has never tracked anything. You set up page tracking and you want to answer the question "What is the probability that if the user visits a product page they will purchase a product?" We could start by artificially making a guess such as Beta(2,8):
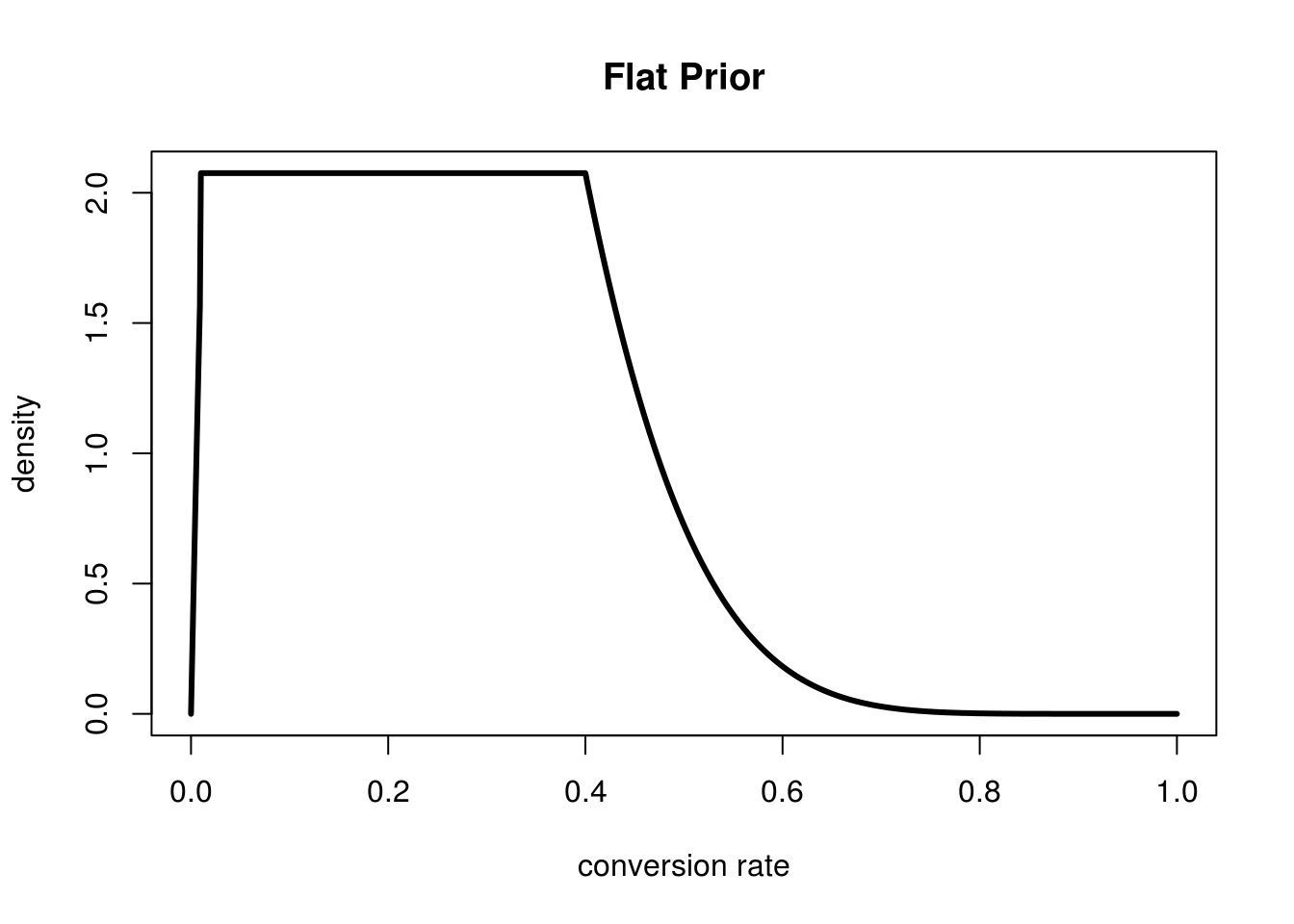
For me personally I feel like that lower tail after 0.4 seems pretty sane, but the peak seems like a complete guess. Between 0.01 and 0.3 I feel that the prior should be uniform. I don't have any extra information and personally don't belief that any one of these is more likely than the other. Here's what I want to see:
This says "I'm pretty skepitical of conversion rates less than 1%, and increasingly skeptical of them as they grow past 40%, everything else I feel equally likely about". The problem here is that I don't have a mathematical equation for this function. I simply used the following code to generate this plot above:
#We need to approximate the dx for this function dx <- 0.001 xs <- seq(0,1,by=dx) #get the values of Beta(2,8) for each dx fxs <- dbeta(xs,2,8) #flatten those values in the range we want flattened prior.ys <- ifelse(xs < 0.01 | xs > 0.4,fxs,dbeta(0.4,2,8)) #normalize so it all sums to 1 prior.ys <- prior.ys/(sum(prior.ys)*dx) plot(xs,prior.ys,type='l',lwd=3,xlab="conversion rate",ylab="density", main="Flat Prior")
The problem with doing this in code is that we no longer have a nice analytical solution for calcuating our normalized Posterior! To demonstrate this let's collect some data. We've installed the tracker and our results show that in the first 12 people 2 converted. Then we have Beta(2,10) for our likelihood. Let's compare our posteriors for our two priors:
Here is the code for calculating each of the Posteriors in the graph above:
# Using the Beta Prior data.alpha <- 2 data.beta <- 10 prior.alpha <- 2 prior.beta <- 8 plot(xs,dbeta(xs,data.alpha+prior.alpha,data.beta+prior.beta),type='l',lwd=3, xlab="conversion rate",ylab="density",main="Comparing Two Posteriors") # Using the Flat Prior data.ys <- dbeta(xs,data.alpha,data.beta) post.ys <- data.ys * prior.ys post.ys <- post.ys/(sum(post.ys)*dx) lines(xs,post.ys,type='l',col="blue",lwd=3) legend(0.7,4,c('Beta(2,8) prior','Flat prior'),c("black","blue"),bty='n')
Because the posterior for the PDF using the Beta distribution is also a Beta distribution any probabilities we want to reason about are easily solved by integration. In R we could use the dbeta function for this. However for our flat prior we don't even know how the Posterior is defined analytically and there is no equivalent to dbeta for this, so how in the world can we integrate over it?
In the post on Monte-Carlo simulations, we learned that it is is possible to approximate integrals by simply sampling from the distributions and calculating the ratio between points in the area we're interested in and points outside. For example to calculate the integral for a Normal distribution with \(\mu=0\) and \(\sigma = 10\) from 3 to 6 we just need this little bit of code:
runs <- 100000 sims <- rnorm(runs,mean=1,sd=10) mc.integral <- sum(sims >= 3 & sims <= 6)/runs
Unfortunately, just like we don't have the equivalent of dbeta we don't have a sampling function like rnorm either. If we had some way to sample from this distribution then we could compute the integral.
We need a Sampler!
Not only would having a method of sampling from our odd Posterior be useful for calculating the Integral, but for a wide range of other tasks, such as comparing two parameter estimates as we did in our A/B testing example. We're going to solve this problem by building what is referred to as a Rejection Sampler.
Looking at the plot of our Posterior we find that we can fit all of our points in a rectangle that ranges from 0.0 to 1.0 on the x-axis and 0 to 4.33 on the y-axis. What we want to do is randomly sample from a uniform distribution on each axis. Then we look at whether or not the point created for these sampled pairs falls under our curve or not. If it is under we accept the sample, otherwise we reject it (hence the name).
For example, we sample two random points A and B on this plane. Point A is (0.4,1.2) and point B is (0.2,2.3), these can be seen here:
We've colored A red because it is outside of our distribution, so we're going to reject it. B however is under the curve so we are going to sample it, which means we're going to take it's 'x' value. In the (x,y) pair x is the value we may or may not be sampling (in this case 0.2) and y determines whether or not we sample it.
Implementation
It is always easy to get the general idea of how an algorithm works, but to really understand it we need to implement it. We'll start by sampling 10,000 points in the space we described:
n.pairs <- 10000 #we'll just put this in a data frame so they're easier to work with xy.pairs <- data.frame(xs=runif(n.pairs,min=0,max=1), ys=runif(n.pairs,min=0,max=4.33))
Now that we have the samples we simply need a function that will take an x and a y then let us know if the point is inside our function or not. There is a slight problem we have to deal with computationally. So far we've been pretending that we are dealing with continuous functions, but the truth is that we've been using discrete approximations (because we are using a computer to calculate everything). In the earlier
The reason this is an issue is that we want to be able to reuse the post.ys that we already calculated. So we only want to sample from values that are accurate to 0.001 (other wise we can't use our sampled xs to look up our index to the ys). To solve
which.nearest <- function(x,xs){ #assumes xs are in order, chooses the first one greater first.bigger <- which(xs > x)[1] #see which of the first largest or the last smallest x is closer too smallest.diff <- which.min(abs(c(xs[first.bigger-1],xs[first.bigger])-x)) #stupid trick to return either the index of the first bigger or the index of the last smaller first.bigger+(smallest.diff-2) }
And now that that detail is out of the way we can go on rejecting/accepting our points!
xy.pairs$post <- sapply(xy.pairs$xs,function(x){ #remember xs is a global we defined ealier post.ys[which.nearest(x,xs)] }) xy.pairs$accept <- xy.pairs$ys < xy.pairs$post
For a sanity check we can plot this all out to make sure our points look right.
One final check we can perform is to plot out the density of our xs and see how it compares to our actual density:
As we can see the empirically observed density of our samples matches up quite well with our Posterior! Now that we have a way to sample from our strange function we can use all the Monte-Carlo techniques we've covered for computing integrals and comparing distributions!
Conclusion
One of the most beautiful things about Monte-Carlo simulations is that they make it relatively easy to model distributions that would be immensely hard to solve analytically. Classical statistics makes heavy use of the Normal Distribution not only because it tends to appear often in nature, but also as an approximating tool. However, there are many distributions found in the real world where the Normal Distribution is a poor approximation. Before the rise of easy access to computing many of these problems had to be ignored or simply solved poorly. With Rejection Sampling and other techniques, we are able to make use of large range of ill-defined distributions that would otherwise be out of our reach!
If you enjoyed this post please subscribe to keep up to date and follow @willkurt!